Genes-, Gene-sets & Features
spata-genes-gene-sets-and-features.RmdGenes and gene-sets are of major importance when it comes to the analysis of every RNA sequencing results. The average count-matrix contains information of these expression levels for more than 20.000 genes. While referring to a few of them by entering their names manually might be tolerable things can get cumbersome rapidly once the number of genes of interest becomes double- or tripple digit. Still the majority of spata-functions needs information of the genes, gene-sets and features of interest in form of character-vectors. The following tutorial introduces important helper functions that make living easier when it comes to working in SPATA and working with these informative variables in general.
# load packages library(SPATA) library(magrittr) # load object spata_obj <- loadSpataObject("data/spata-obj-genes-features-example.RDS")
Genes and gene-sets
Setting up a gene-set data.frame
The term gene-set refers to a collection of several genes that are known to interact with each other or being co-expressed together under certain conditions. Information about the gene-sets defined in your spata-object is stored in the slot @used_genesets which contains a data.frame of two character-variables.
- ont: The name of the gene-sets.
- gene: The belonging genes.
In SPATA gene-set names are functionally divided into their class and their actual name. The class is determined by the character string that comes before the first *_*, the name refers to the rest. Subsequent chapters will highlight why this functional separation is useful as well as why it is useful to really think about the way you name your personally defined gene-sets. In order to obtain an overview of your saved gene-sets run getGeneSetOverview().
getGeneSetOverview(spata_obj)
By default initiateSpataObject() assigns a gene-set data.frame to your spata-object containing a variety of predefined gene-sets from the Molecular Signature Database. Run data(gsdf) to load and ?gsdf to get information about it and what the class-names stand for. If you want to add new ones you can do that via addGeneSet().
# add a new gene-set of three genes spata_obj <- addGeneSet(object = spata_obj, class_name = "New", gs_name = "example_1", genes = c('SLC35E2A', 'NADK', 'GFAP')) # genes that exist in your expression matrix getGeneSetOverview(spata_obj)
A more convenient way would be to open an interactive application via addGeneSetsInteracive() as displayed below. Make sure to close the application via the respective button and to store the resulting object in the same object in order to overwrite it and not to overcrowd your global environment with spata-objects that only differ in a few gene-sets.
spata_obj <- addGeneSetsInteractive(spata_obj)
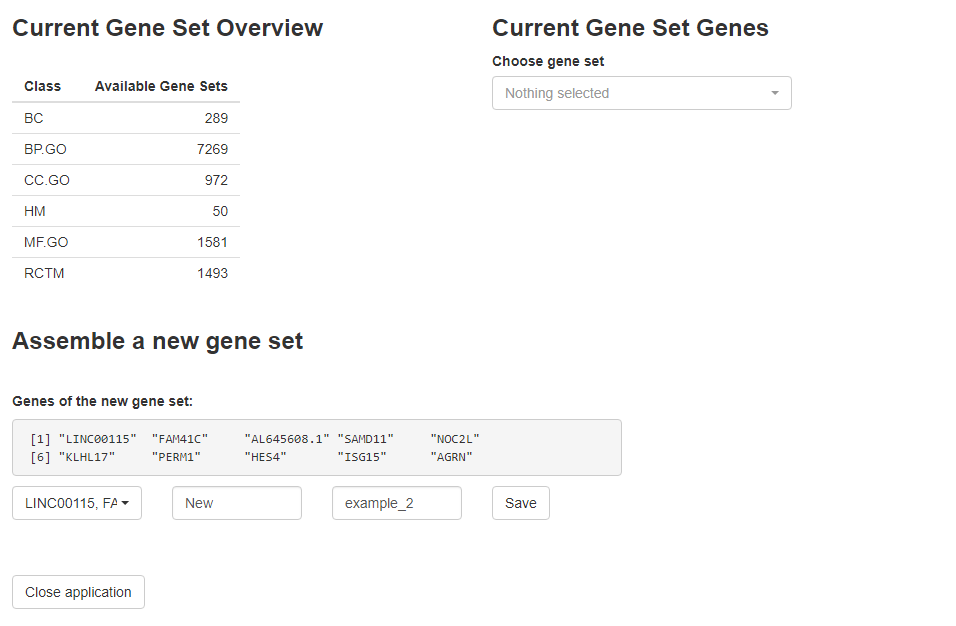
Figure 1. Interface of addGeneSetsInteractive()
The gene-set data.frame is infinitely expandable. If you want to discard gene-sets or alter them you can either use addGeneSet() with argument overwrite set to TRUE in order to overwrite existing ones or discardGeneSets() as shown below.
# discard gene-sets by name spata_obj <- discardGeneSets(object = spata_obj, gs_names = c("New_example_2")) # display updated overview getGeneSetOverview(spata_obj)
In order to save the gene-set data.frame of one particular spata-object for further use with other spata-objects use saveGeneSetDf() which saves the @used_genesets slot of the specified object as a .RDS-file.
saveGeneSetDf(object = spata_obj, output_path = "/your/path/to/storage-folder", filename = "gene-set-df.RDS") # for example
Adjusting a gene-set data.frame
Given the nature of your sample it will happen that your expression matrix does not contain genes (0 reads across all barcodes) although they appear in certain gene-sets. joinWithGeneSets() which works under the hood of a variety of functions does not allow for gene-sets to join your data if less than 25% of the genes needed to form that gene-set are found. If this threshold is to loose to you you can use adjustGeneSetDf() to set the limit yourself. It will then discard all gene-sets whoose belonging genes are found to a percentage lower than the threshold you set. (Discarding gene-sets from your gene-set data.frame effectively makes it impossible to work with them within SPATA.)
Referring to genes and gene-sets
getGenes() and getGeneSets() return character vectors which are valid inputs for all SPATA-functions in which one has to refer to genes or gene-sets in any way.
Obtaining gene-set names
As we have seen above gene-sets are a set of genes that are functionally gathered under a certain name. getGeneSets() does not return the genes of certain gene-sets but the gene-set-names as strings It does that either as a single character vector or as a named list.
# to get all gene-set names set 'of_class' to 'all' all_gs <- getGeneSets(spata_obj, of_class = "all") length(all_gs)
## [1] 11655head(all_gs, 10)
## [1] "BC_RELA_PATHWAY" "BC_NO1_PATHWAY"
## [3] "BC_CSK_PATHWAY" "BC_SRCTMRPTP_PATHWAY"
## [5] "BC_AMI_PATHWAY" "BC_GRANULOCYTES_PATHWAY"
## [7] "BC_LYM_PATHWAY" "BC_ARAP_PATHWAY"
## [9] "BC_AGR_PATHWAY" "BC_AKAP95_PATHWAY"# get gene-set names of certain classes getGeneSets(spata_obj, of_class = c("HM", "New"), simplify = TRUE # as a single vector ) %>% tail() # show last six
## [1] "HM_ALLOGRAFT_REJECTION" "HM_SPERMATOGENESIS" "HM_KRAS_SIGNALING_UP"
## [4] "HM_KRAS_SIGNALING_DN" "HM_PANCREAS_BETA_CELLS" "New_example_1"# get gene-set names of certain classes getGeneSets(spata_obj, of_class = c("HM", "New"), simplify = FALSE # as a named list ) %>% lapply(head) #show first six
## $HM
## [1] "HM_TNFA_SIGNALING_VIA_NFKB" "HM_HYPOXIA"
## [3] "HM_CHOLESTEROL_HOMEOSTASIS" "HM_MITOTIC_SPINDLE"
## [5] "HM_WNT_BETA_CATENIN_SIGNALING" "HM_TGF_BETA_SIGNALING"
##
## $New
## [1] "New_example_1"To subset your gene-sets of interest additionally make use of the argument index which takes a character-string as a regular expression and filters the gene-sets to be returned again according to it.
# get all gene-sets of class HALLMARK referring to something specific getGeneSets(spata_obj, of_class = "HM", index = "ESTROGEN|ANDROGEN")
## [1] "HM_ESTROGEN_RESPONSE_EARLY" "HM_ESTROGEN_RESPONSE_LATE"
## [3] "HM_ANDROGEN_RESPONSE"If you want to skim your gene-sets interactively use getGeneSetsInteractive() which opens an application in your R-Studio’s viewer pane and returns a vector of chosen gene-sets.
Obtaining gene names
Eventually expression matrices contain information about gene expression levels and not gene-set expression levels. If you are looking for genes of certain gene-sets getGenes() provides an easy we to do just that.
# getGenes() without any further specification will return all genes found # in your expression matrix ... all_genes <- getGenes(spata_obj) # ... which can be quite a lot length(all_genes)
## [1] 21441# with regards to gene-sets it makes more sense to break genes down by # their gene-set belonging estrogen_gs <- getGeneSets(spata_obj, of_class = "HM", index = "ESTROGEN") estrogen_genes <- getGenes(spata_obj, of_gene_sets = estrogen_gs, simplify = FALSE) # to return them sorted in a list #hint: "..." was added manually for visualization purpose as these gene-sets contain # more than 150 genes
## $HM_ESTROGEN_RESPONSE_EARLY
## [1] "GREB1" "CA12" "SLC9A3R1" "MYB" "ANXA9" "IGFBP4"
## [7] "SYBU" "NPY1R" "PDZK1" "NRIP1" "MLPH" "HSPB8"
## [13] "EGR3" "KRT19" "LRIG1" "KDM4B" "PGR" "RHOBTB3"
## [19] "TPD52L1" "ELOVL2" "..."
##
## $HM_ESTROGEN_RESPONSE_LATE
## [1] "SLC9A3R1" "TPD52L1" "PRSS23" "CA12" "PDZK1" "ANXA9"
## [7] "CELSR2" "PGR" "RET" "MYB" "TPBG" "EGR3"
## [13] "ARL3" "OLFM1" "NPY1R" "SCNN1A" "XBP1" "AREG"
## [19] "IL17RB" "NRIP1" "..."If you want to work with genes manually and skim them interactively use getGenesInteractive() which opens an application in your R-Studio’s viewer pane.
Features
In SPATA features refer to all kinds of information of barcode-spots that do not fall explicitly under gene- or gene-set expression levels. This includes in particular group belonging such as clustering or spatial segmentation. But additional numeric features such as monocle3-pseudotime or everything else that has been computed is considered to be a feature as well. Feature information is stored - in a tidy data fashion - in the slot @fdata. To obtain all feature information use getFeatureData().
getFeatureData(object = spata_obj)
In order to specify features of interest as input in functions you need to specify the name that particular feature variable carries. Several functions only work with feature data of one kind - either numeric features or discrete/categorical features. The function getFeatureNames() provides you with information about all currently available features in your spata-object as well as about the class they belong to.
# get all feature names getFeatureNames(object = spata_obj)
## numeric integer numeric numeric
## "nCount_RNA" "nFeature_RNA" "percent.mt" "percent.RB"
## factor factor character character
## "RNA_snn_res.0.8" "seurat_clusters" "segment" "igraph_clusters"# get only names of numeric features numeric_features <- getFeatureNames(object = spata_obj, of_class = c("numeric")) # output numeric_features
## numeric numeric numeric
## "nCount_RNA" "percent.mt" "percent.RB"# get only names of discrete features discrete_features <- getFeatureNames(object = spata_obj, of_class = c("character", "factor")) # output discrete_features
## factor factor character character
## "RNA_snn_res.0.8" "seurat_clusters" "segment" "igraph_clusters"Additionally getFeatureValues() allows to quickly access the unique values of discrete feature variables such as clusters.
getFeatureValues(spata_obj, features = discrete_features)
## $RNA_snn_res.0.8
## [1] 2 4 1 3 0 5 6 7
## Levels: 0 1 2 3 4 5 6 7
##
## $seurat_clusters
## [1] 2 4 1 3 0 5 6 7
## Levels: 0 1 2 3 4 5 6 7
##
## $segment
## [1] ""
##
## $igraph_clusters
## [1] "Cluster 5" "Cluster 1" "Cluster 6" "Cluster 3" "Cluster 2" "Cluster 7"
## [7] "Cluster 4"Whenever a warning message appears telling you that a certain gene, gene-set or feature was not found this means that the character string provided was not found throughout your spata-object. Check for typos in this case. The same is true for gene-set names with regards to the ont-variable in the gene-set data.frame.
Example
The output vectors of getGenes(), getGeneSets() and getFeatureNames() are a perfectly valid inputs for arguments like color_to or variables.
hallmark_gs <- getGeneSets(object = spata_obj, of_class = "HM") # output hallmark_gs[1:10]
## [1] "HM_TNFA_SIGNALING_VIA_NFKB" "HM_HYPOXIA"
## [3] "HM_CHOLESTEROL_HOMEOSTASIS" "HM_MITOTIC_SPINDLE"
## [5] "HM_WNT_BETA_CATENIN_SIGNALING" "HM_TGF_BETA_SIGNALING"
## [7] "HM_IL6_JAK_STAT3_SIGNALING" "HM_DNA_REPAIR"
## [9] "HM_G2M_CHECKPOINT" "HM_APOPTOSIS"# create a spata-data.frame joined_df <- joinWith(object = spata_obj, spata_df = getCoordinates(spata_obj), gene_sets = hallmark_gs, features = discrete_features) #output joined_df
With our vector hallmark_gs containing our gene-sets of interest we can work throughout our R-session.
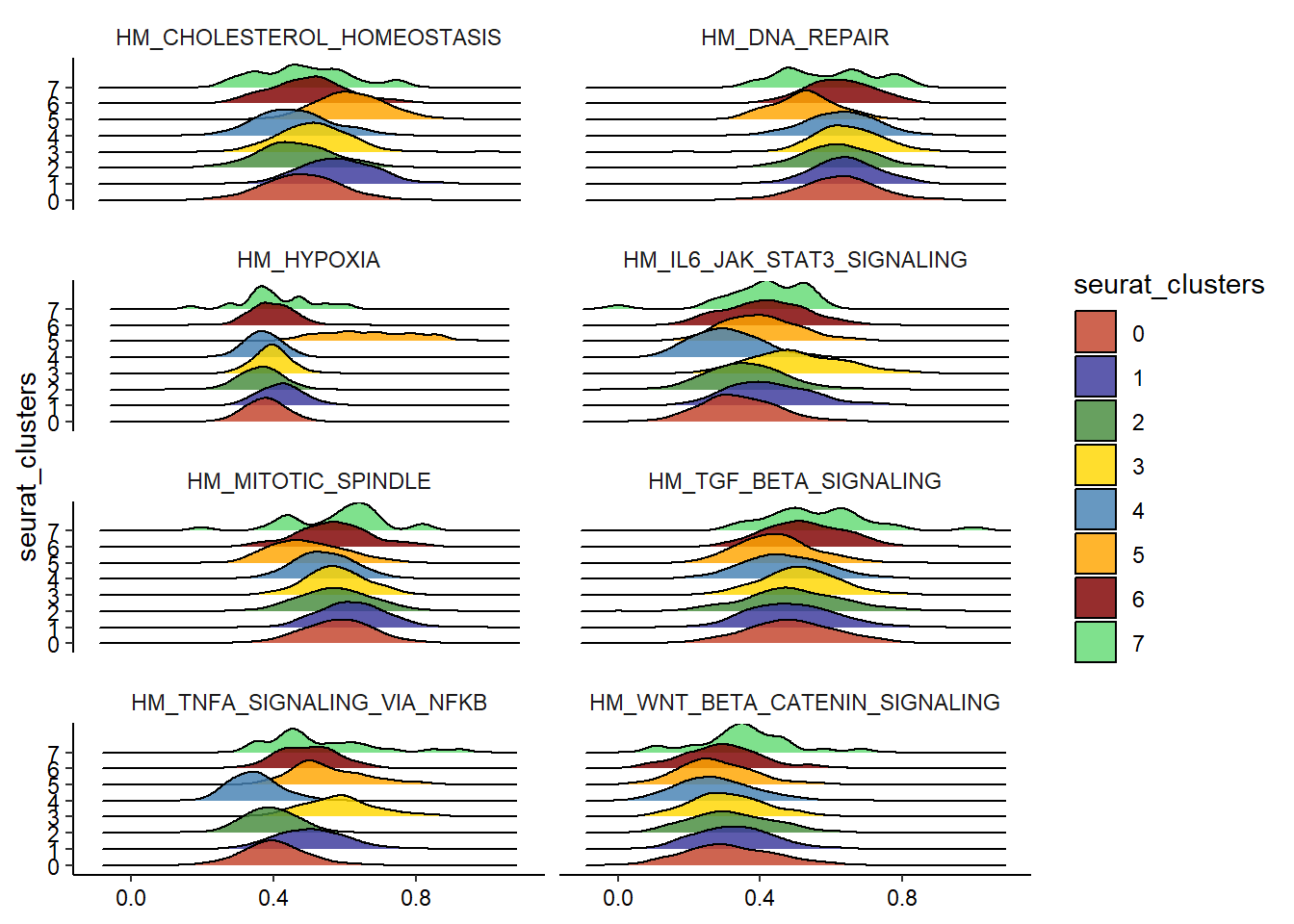
plotDistributionAcross2(df = joined_df, variables = hallmark_gs[1:8], across = "seurat_clusters", plot_type = "ridgeplot", ncol = 2)